
FMODB ID: J3LZ9
Calculation Name: 4FKS-AB-MD340-md3_14ns
Preferred Name: Cyclin-dependent kinase 2
Target Type: SINGLE PROTEIN
Ligand Name: n-[(4-{[(z)-(7-oxo-6,7-dihydro-8h-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}phenyl)sulfonyl]acetamide | acetyl group
Ligand 3-letter code: 46K | ACE
Ligand of Interest (LOI): LIG
PDB ID: 4FKS
Chain ID: AB
ChEMBL ID: CHEMBL301
UniProt ID: P24941
Base Structure: MD
Registration Date: 2022-09-07
Reference: K. Takaba et al., Protein-ligand binding affinity prediction of cyclin-dependent kinase-2 inhibitors by dynamically averaged fragment molecular orbital-based interaction energy, J Comput Chem. 2022;43:1362-1371.
DOI: 10.1002/jcc.26940
Apendix: None
Modeling method
| Optimization | MOE:Amber10:EHT |
|---|---|
| Restraint | OptHLSolv |
| Protonation | BaseStructure_original |
| Complement | BaseStructure_original |
| Water | Water molecules within 6 angstrom of a protein-ligand complex. |
| Procedure | Manual calculation |
FMO calculation
| FMO method | FMO2-MP2/6-31G(d) |
|---|---|
| Fragmentation | Auto |
| Number of fragment | 2696 |
| LigandResidueName | LIG |
| LigandFragmentNumber | 299 |
| LigandCharge | |
| Software | MIZUHO/ABINIT-MP 4.0 |
Total energy (hartree)
| FMO2-HF: Electronic energy | -18957174.107788 |
|---|---|
| FMO2-HF: Nuclear repulsion | 18655415.601617 |
| FMO2-HF: Total energy | -301758.506171 |
| FMO2-MP2: Total energy | -302583.417011 |
3D Structure
Ligand structure
LIG
Ligand Interaction
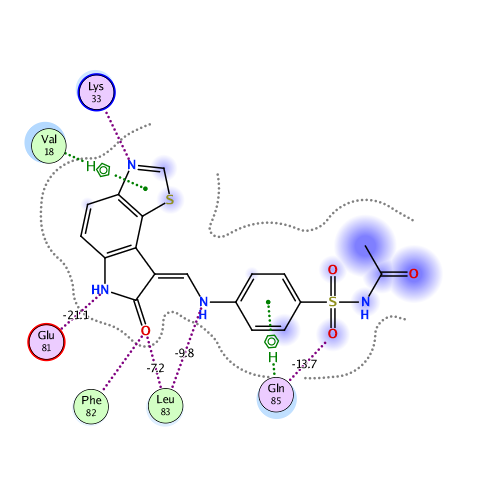
Ligand binding energy (frag 299 : frag 1-298)
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| -105.330889 | -80.557851 | 70.305563 | -28.667328 | -66.411273 | -0.032229 |
Interactive mode: IFIE and PIEDA for fragment #299(A:299:LIG)
Summations of interaction energy for
fragment #299(A:299:LIG)
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
Interaction energy analysis for fragmet #299(A:299:LIG)
| frag_NumFragment number. | ChainChain species. | Res #Residue number. | RES3-letter code of amino acid residue, ligand and solvent molecule. | FCHARGEFormal charge [e]. | q_MullikenFragment charge evaluated by Mulliken charge [e]. | q_NPAFragment charge evaluated by natural population analysis(NPA) charge [e]. | DISTDistance from Ligand [Å]. | TotalIFIE at MP2 level [kcal/mol]. | ESElectro static interaction energy by PIEDA [kcal/mol]. | EXExchange-repulsion energy by PIEDA [kcal/mol]. | CT+mixCharge transfer and mixing terms energy by PIEDA [kcal/mol]. | DI(MP2)Dispersion energy by PIEDA [kcal/mol]. | q(I=>J)Charge transfer value from I to J fragmens [e]. |
|---|