
FMODB ID: N3YQQ
Calculation Name: 7KVR-B-Xray89
Preferred Name:
Target Type:
Ligand Name: n~4~,n~4~-dimethylpyridine-2,4-diamine | di(hydroxyethyl)ether | dimethyl sulfoxide
Ligand 3-letter code: X4V | PEG | DMS
Ligand of Interest (LOI): X4V
PDB ID: 7KVR
Chain ID: B
Base Structure: X-ray
Registration Date: 2021-02-22
Reference: T. Ohyama, K. Kamisaka, C. Watanabe, T. Honma et. al., FMO-based interaction energy analysis of SARS-Cov-2 main protease and ligand complexes, To be published.
Apendix: None
Modeling method
| Optimization | MOE:Amber10:EHT |
|---|---|
| Restraint | OptHLSideSolv |
| Protonation | MOE:Protonate 3D |
| Complement | MOE:Structure Preparation |
| Water | Waters within average of the temperature factors of receptor. |
| Procedure | Auto-FMO protocol ver. 1.20200406 |
FMO calculation
| FMO method | FMO2-MP2/6-31G(d) |
|---|---|
| Fragmentation | Auto |
| Number of fragment | 50 |
| LigandResidueName | X4V |
| LigandFragmentNumber | 3 |
| LigandCharge | X4V=1 |
| Software | MIZUHO/ABINIT-MP 4.0(SMP) |
Total energy (hartree)
| FMO2-HF: Electronic energy | -17371.216798 |
|---|---|
| FMO2-HF: Nuclear repulsion | 11119.817593 |
| FMO2-HF: Total energy | -6251.399205 |
| FMO2-MP2: Total energy | -6265.18415 |
3D Structure
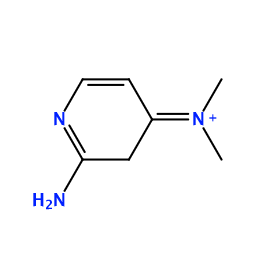
Ligand structure
X4V
Ligand Interaction
Ligand binding energy
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| 10.944 | 19.902 | 12.695 | -5.004 | -16.650 | 0.036 |
Interactive mode: IFIE and PIEDA for fragment #3(B:703:X4V )
Summations of interaction energy for
fragment #3(B:703:X4V )
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| 10.944 | 19.902 | 12.695 | -5.004 | -16.65 | -0.036 |
Interaction energy analysis for fragmet #3(B:703:X4V )
| frag_NumFragment number. | ChainChain species. | Res #Residue number. | RES3-letter code of amino acid residue, ligand and solvent molecule. | FCHARGEFormal charge [e]. | q_MullikenFragment charge evaluated by Mulliken charge [e]. | q_NPAFragment charge evaluated by natural population analysis(NPA) charge [e]. | DISTDistance from Ligand [Å]. | TotalIFIE at MP2 level [kcal/mol]. | ESElectro static interaction energy by PIEDA [kcal/mol]. | EXExchange-repulsion energy by PIEDA [kcal/mol]. | CT+mixCharge transfer and mixing terms energy by PIEDA [kcal/mol]. | DI(MP2)Dispersion energy by PIEDA [kcal/mol]. | q(I=>J)Charge transfer value from I to J fragmens [e]. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B | 701 | PEG | 0 | 0.000 | 0.000 | 36.194 | 0.070 | 0.070 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | B | 702 | DMS | 0 | 0.037 | 0.035 | 27.420 | 0.256 | 0.256 | 0.000 | 0.000 | 0.000 | 0.000 |
| 4 | B | 704 | DMS | 0 | -0.013 | -0.003 | 25.490 | 0.069 | 0.069 | 0.000 | 0.000 | 0.000 | 0.000 |
| 5 | B | 705 | DMS | 0 | 0.000 | 0.000 | 20.497 | -0.437 | -0.437 | 0.000 | 0.000 | 0.000 | 0.000 |
| 6 | B | 801 | HOH | 0 | 0.000 | 0.000 | 27.515 | -0.165 | -0.165 | 0.000 | 0.000 | 0.000 | 0.000 |
| 7 | B | 802 | HOH | 0 | 0.025 | 0.021 | 42.631 | 0.050 | 0.050 | 0.000 | 0.000 | 0.000 | 0.000 |
| 8 | B | 803 | HOH | 0 | -0.029 | -0.023 | 43.021 | -0.029 | -0.029 | 0.000 | 0.000 | 0.000 | 0.000 |
| 9 | B | 804 | HOH | 0 | 0.015 | 0.004 | 2.508 | -9.149 | -8.063 | 1.687 | -1.101 | -1.672 | -0.015 |
| 10 | B | 805 | HOH | 0 | -0.012 | -0.017 | 2.587 | -9.336 | -8.385 | 1.439 | -1.009 | -1.380 | -0.014 |
| 11 | B | 806 | HOH | 0 | -0.017 | -0.019 | 26.174 | -0.185 | -0.185 | 0.000 | 0.000 | 0.000 | 0.000 |
| 12 | B | 807 | HOH | 0 | 0.000 | 0.000 | 29.345 | -0.018 | -0.018 | 0.000 | 0.000 | 0.000 | 0.000 |
| 13 | B | 808 | HOH | 0 | 0.038 | 0.027 | 3.886 | -3.404 | -3.262 | 0.003 | -0.046 | -0.100 | 0.000 |
| 14 | B | 809 | HOH | 0 | 0.000 | 0.000 | 18.246 | -0.050 | -0.050 | 0.000 | 0.000 | 0.000 | 0.000 |
| 15 | B | 810 | HOH | 0 | 0.000 | 0.000 | 22.694 | -0.182 | -0.182 | 0.000 | 0.000 | 0.000 | 0.000 |
| 16 | B | 811 | HOH | 0 | 0.023 | 0.021 | 21.477 | 0.253 | 0.253 | 0.000 | 0.000 | 0.000 | 0.000 |
| 17 | B | 812 | HOH | 0 | 0.003 | 0.002 | 43.935 | -0.029 | -0.029 | 0.000 | 0.000 | 0.000 | 0.000 |
| 18 | B | 813 | HOH | 0 | 0.004 | 0.004 | 22.212 | 0.019 | 0.019 | 0.000 | 0.000 | 0.000 | 0.000 |
| 19 | B | 814 | HOH | 0 | 0.000 | 0.000 | 16.389 | -0.358 | -0.358 | 0.000 | 0.000 | 0.000 | 0.000 |
| 20 | B | 815 | HOH | 0 | 0.000 | 0.000 | 13.303 | -0.440 | -0.440 | 0.000 | 0.000 | 0.000 | 0.000 |
| 21 | B | 816 | HOH | 0 | 0.000 | 0.000 | 29.620 | -0.088 | -0.088 | 0.000 | 0.000 | 0.000 | 0.000 |
| 22 | B | 817 | HOH | 0 | 0.026 | 0.022 | 8.951 | -0.266 | -0.266 | 0.000 | 0.000 | 0.000 | 0.000 |
| 23 | B | 818 | HOH | 0 | 0.023 | 0.019 | 18.112 | -0.015 | -0.015 | 0.000 | 0.000 | 0.000 | 0.000 |
| 24 | B | 819 | HOH | 0 | -0.037 | -0.035 | 31.966 | 0.044 | 0.044 | 0.000 | 0.000 | 0.000 | 0.000 |
| 25 | B | 820 | HOH | 0 | 0.025 | 0.024 | 15.479 | 0.374 | 0.374 | 0.000 | 0.000 | 0.000 | 0.000 |
| 26 | B | 821 | HOH | 0 | 0.023 | 0.021 | 36.823 | -0.042 | -0.042 | 0.000 | 0.000 | 0.000 | 0.000 |
| 27 | B | 822 | HOH | 0 | -0.006 | -0.005 | 9.833 | -0.552 | -0.552 | 0.000 | 0.000 | 0.000 | 0.000 |
| 28 | B | 823 | HOH | 0 | 0.000 | 0.000 | 10.429 | -0.967 | -0.967 | 0.000 | 0.000 | 0.000 | 0.000 |
| 29 | B | 824 | HOH | 0 | 0.000 | 0.000 | 11.616 | -0.282 | -0.282 | 0.000 | 0.000 | 0.000 | 0.000 |
| 30 | B | 825 | HOH | 0 | 0.000 | 0.000 | 22.251 | -0.207 | -0.207 | 0.000 | 0.000 | 0.000 | 0.000 |
| 31 | B | 826 | HOH | 0 | 0.026 | 0.021 | 4.996 | -3.386 | -3.359 | 0.000 | -0.001 | -0.027 | 0.000 |
| 32 | B | 827 | HOH | 0 | 0.000 | 0.000 | 16.056 | -0.357 | -0.357 | 0.000 | 0.000 | 0.000 | 0.000 |
| 33 | B | 828 | HOH | 0 | 0.000 | 0.000 | 18.839 | -0.309 | -0.309 | 0.000 | 0.000 | 0.000 | 0.000 |
| 34 | B | 829 | HOH | 0 | 0.030 | 0.022 | 28.717 | -0.157 | -0.157 | 0.000 | 0.000 | 0.000 | 0.000 |
| 35 | B | 830 | HOH | 0 | 0.004 | 0.003 | 18.117 | 0.300 | 0.300 | 0.000 | 0.000 | 0.000 | 0.000 |
| 36 | B | 831 | HOH | 0 | 0.000 | 0.000 | 11.127 | -0.725 | -0.725 | 0.000 | 0.000 | 0.000 | 0.000 |
| 37 | B | 832 | HOH | 0 | 0.006 | 0.007 | 9.877 | 0.584 | 0.584 | 0.000 | 0.000 | 0.000 | 0.000 |
| 38 | B | 833 | HOH | 0 | -0.003 | -0.003 | 22.287 | -0.221 | -0.221 | 0.000 | 0.000 | 0.000 | 0.000 |
| 39 | B | 834 | HOH | 0 | -0.023 | -0.021 | 35.876 | 0.045 | 0.045 | 0.000 | 0.000 | 0.000 | 0.000 |
| 40 | B | 836 | HOH | 0 | 0.000 | 0.000 | 12.845 | -0.601 | -0.601 | 0.000 | 0.000 | 0.000 | 0.000 |
| 41 | B | 837 | HOH | 0 | -0.001 | -0.001 | 20.326 | 0.139 | 0.139 | 0.000 | 0.000 | 0.000 | 0.000 |
| 42 | B | 838 | HOH | 0 | -0.023 | -0.021 | 23.912 | 0.164 | 0.164 | 0.000 | 0.000 | 0.000 | 0.000 |
| 43 | B | 839 | HOH | 0 | 0.024 | 0.022 | 25.259 | -0.196 | -0.196 | 0.000 | 0.000 | 0.000 | 0.000 |
| 44 | B | 840 | HOH | 0 | -0.026 | -0.022 | 7.468 | -1.722 | -1.722 | 0.000 | 0.000 | 0.000 | 0.000 |
| 45 | B | 841 | HOH | 0 | 0.001 | -0.002 | 17.099 | -0.038 | -0.038 | 0.000 | 0.000 | 0.000 | 0.000 |
| 46 | B | 842 | HOH | 0 | -0.027 | -0.022 | 20.505 | 0.252 | 0.252 | 0.000 | 0.000 | 0.000 | 0.000 |
| 47 | B | 843 | HOH | 0 | -0.024 | -0.022 | 22.593 | -0.178 | -0.178 | 0.000 | 0.000 | 0.000 | 0.000 |
| 48 | B | 844 | HOH | 0 | -0.027 | -0.022 | 16.471 | -0.427 | -0.427 | 0.000 | 0.000 | 0.000 | 0.000 |
| 49 | B | 845 | HOH | 0 | -0.001 | -0.002 | 10.781 | -0.215 | -0.215 | 0.000 | 0.000 | 0.000 | 0.000 |
| 50 | B | 703 | X4V | 1 | 0.969 | 0.978 | 2.393 | 43.058 | 49.810 | 9.566 | -2.847 | -13.471 | -0.007 |