
FMODB ID: R58L8
Calculation Name: 2V63-ABIJ-Xray434
Preferred Name:
Target Type:
Ligand Name: 2-carboxyarabinitol-1,5-diphosphate | lysine nz-carboxylic acid | 4-hydroxyproline | n-methyl methionine | s-methylcysteine | 1,2-ethanediol
Ligand 3-letter code: CAP | KCX | HYP | MME | SMC | EDO
Ligand of Interest (LOI): CAP
PDB ID: 2V63
Chain ID: ABIJ
UniProt ID: P00877
Base Structure: X-ray
Registration Date: 2023-09-26
Reference: M. Fujii, S. Tanaka, Interspecies Comparison of Interaction Energies between Photosynthetic Protein RuBisCO and 2CABP Ligand, Int. J. Mol. Sci. 2022, 23(19), 11347.
Apendix: None
Modeling method
| Optimization | MOE:Amber10:EHT |
|---|---|
| Restraint | OptH |
| Protonation | MOE:Protonate 3D |
| Complement | MOE:Structure Preparation |
| Water | No |
| Procedure | Manual calculation |
FMO calculation
| FMO method | FMO2-MP2/6-31G(d) |
|---|---|
| Fragmentation | Manual |
| Number of fragment | 1215 |
| LigandResidueName | CAP |
| LigandFragmentNumber | 1214 |
| LigandCharge | |
| Software | ABINIT-MP - Open Ver. 1 Rev. 22 / 20200603(SMP) |
Total energy (hartree)
| FMO2-HF: Electronic energy | -43814885.785359 |
|---|---|
| FMO2-HF: Nuclear repulsion | 43326549.465638 |
| FMO2-HF: Total energy | -488336.31972 |
| FMO2-MP2: Total energy | -489729.556147 |
3D Structure
Ligand structure
CAP
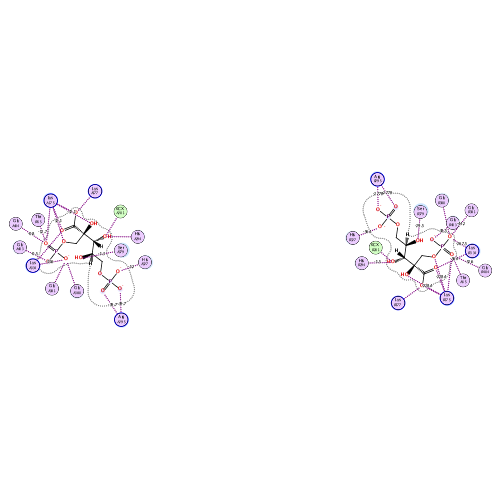
Ligand Interaction
Ligand binding energy
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| -889.778 | -902.516 | 185.881 | -89.393 | -83.749 | -0.718 |
Interactive mode: IFIE and PIEDA for fragment #1214(A:476:CAP)
Summations of interaction energy for
fragment #1214(A:476:CAP)
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
Interaction energy analysis for fragmet #1214(A:476:CAP)
| frag_NumFragment number. | ChainChain species. | Res #Residue number. | RES3-letter code of amino acid residue, ligand and solvent molecule. | FCHARGEFormal charge [e]. | q_MullikenFragment charge evaluated by Mulliken charge [e]. | q_NPAFragment charge evaluated by natural population analysis(NPA) charge [e]. | DISTDistance from Ligand [Å]. | TotalIFIE at MP2 level [kcal/mol]. | ESElectro static interaction energy by PIEDA [kcal/mol]. | EXExchange-repulsion energy by PIEDA [kcal/mol]. | CT+mixCharge transfer and mixing terms energy by PIEDA [kcal/mol]. | DI(MP2)Dispersion energy by PIEDA [kcal/mol]. | q(I=>J)Charge transfer value from I to J fragmens [e]. |
|---|