
FMODB ID: XVQQ1
Calculation Name: 1DB1-A-Xray14
Preferred Name: Vitamin D receptor
Target Type: SINGLE PROTEIN
Ligand Name: 5-{2-[1-(5-hydroxy-1,5-dimethyl-hexyl)-7a-methyl-octahydro-inden-4-ylidene]-ethylidene}-4-methylene-cyclohexane-1,3-diol
ligand 3-letter code: VDX
PDB ID: 1DB1
Chain ID: A
ChEMBL ID: CHEMBL1977
UniProt ID: P11473
Base Structure: X-ray
Registration Date: 2018-03-13
Reference:
Apendix: None
Modeling method
| Optimization | Other |
|---|---|
| Restraint | Other |
| Protonation | Based on pKa values evaluated by PROPKA |
| Complement | SWISS Model |
| Water | Other |
| Procedure | Manual calculation |
FMO calculation
| FMO method | FMO2-MP2/6-31G(d) |
|---|---|
| Fragmentation | Auto |
| Number of fragment | 419 |
| LigandResidueName | VDX |
| LigandFragmentNumber | 424 |
| LigandCharge | VDX=0 |
| Software | MIZUHO/ABINIT-MP 4.0(SMP) |
Total energy (hartree)
| FMO2-HF: Electronic energy | -4228259.681873 |
|---|---|
| FMO2-HF: Nuclear repulsion | 4112107.791054 |
| FMO2-HF: Total energy | -116151.890818 |
| FMO2-MP2: Total energy | -116481.74157 |
3D Structure
Ligand structure
VDX
Ligand Interaction
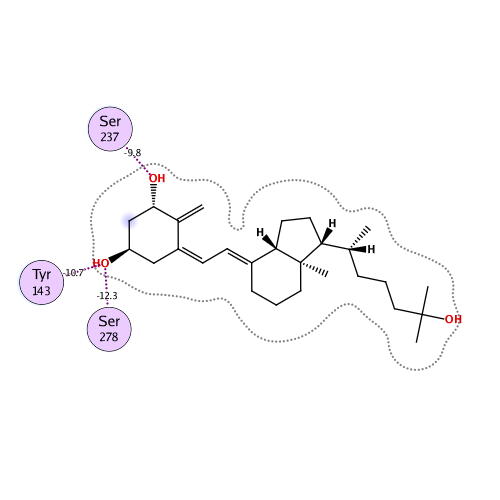
Ligand binding energy
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| -128.345 | -93.584 | 92.641 | -36.401 | -91.003 | 0.042 |
Interactive mode: IFIE and PIEDA for fragment #424(::)
Summations of interaction energy for
fragment #424(::)
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
Interaction energy analysis for fragmet #424(::)
| frag_NumFragment number. | ChainChain species. | Res #Residue number. | RES3-letter code of amino acid residue, ligand and solvent molecule. | FCHARGEFormal charge [e]. | q_MullikenFragment charge evaluated by Mulliken charge [e]. | q_NPAFragment charge evaluated by natural population analysis(NPA) charge [e]. | DISTDistance from Ligand [Å]. | TotalIFIE at MP2 level [kcal/mol]. | ESElectro static interaction energy by PIEDA [kcal/mol]. | EXExchange-repulsion energy by PIEDA [kcal/mol]. | CT+mixCharge transfer and mixing terms energy by PIEDA [kcal/mol]. | DI(MP2)Dispersion energy by PIEDA [kcal/mol]. | q(I=>J)Charge transfer value from I to J fragmens [e]. |
|---|