
FMODB ID: Z2JRN
Calculation Name: 7C33-D-Xray89
Preferred Name:
Target Type:
Ligand Name: adenosine-5-diphosphoribose
ligand 3-letter code: APR
PDB ID: 7C33
Chain ID: D
Base Structure: X-ray
Registration Date: 2021-03-24
Reference: T. Ohyama, K. Kamisaka, C. Watanabe, T. Honma et. al., FMO-based interaction energy analysis of SARS-Cov-2 related protein and ligand complexes, To be published.
Apendix: None
Modeling method
| Optimization | MOE:Amber10:EHT |
|---|---|
| Restraint | OptHLSideSolv |
| Protonation | MOE:Protonate 3D |
| Complement | MOE:Structure Preparation |
| Water | Waters within average of the temperature factors of receptor. |
| Procedure | Auto-FMO protocol ver. 1.20200406 |
FMO calculation
| FMO method | FMO2-MP2/6-31G(d) |
|---|---|
| Fragmentation | Auto |
| Number of fragment | 166 |
| LigandResidueName | APR |
| LigandFragmentNumber | 166 |
| LigandCharge | APR=-2 |
| Software | ABINIT-MP - Open Ver.1 Rev. 16 beta / 20190613 |
Total energy (hartree)
| FMO2-HF: Electronic energy | -1715935.548883 |
|---|---|
| FMO2-HF: Nuclear repulsion | 1651512.921026 |
| FMO2-HF: Total energy | -64422.627856 |
| FMO2-MP2: Total energy | -64610.281234 |
3D Structure
Ligand structure
APR
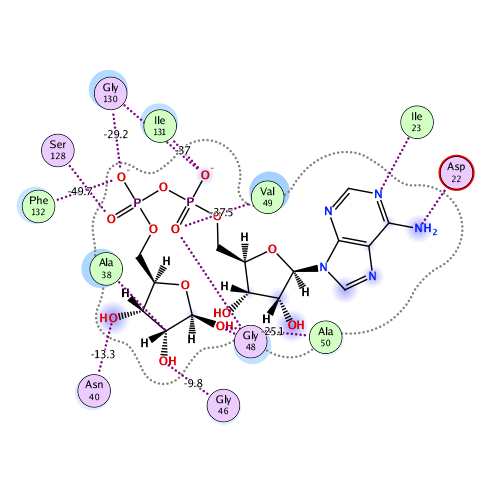
Ligand Interaction
Ligand binding energy
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| -420.226 | -396.670 | 149.778 | -58.751 | -114.586 | -0.333 |
Interactive mode: IFIE and PIEDA for fragment #166(D:201:APR )
Summations of interaction energy for
fragment #166(D:201:APR )
| IFIE [kcal/mol] | PIEDA [kcal/mol] | Charge transfer value [e] | |||
|---|---|---|---|---|---|
| IFIE SUMIFIE SUM at MP2 level. | ESElectro static interaction energy. | EXExchange-repulsion energy. | CT+mixCharge transfer and mixing terms energy. | DI(MP2)Dispersion energy. | q(I=>J)Charge transfer value from I to J fragments. |
| -420.226 | -396.67 | 149.778 | -58.751 | -114.586 | 0.333 |
Interaction energy analysis for fragmet #166(D:201:APR )
| frag_NumFragment number. | ChainChain species. | Res #Residue number. | RES3-letter code of amino acid residue, ligand and solvent molecule. | FCHARGEFormal charge [e]. | q_MullikenFragment charge evaluated by Mulliken charge [e]. | q_NPAFragment charge evaluated by natural population analysis(NPA) charge [e]. | DISTDistance from Ligand [Å]. | TotalIFIE at MP2 level [kcal/mol]. | ESElectro static interaction energy by PIEDA [kcal/mol]. | EXExchange-repulsion energy by PIEDA [kcal/mol]. | CT+mixCharge transfer and mixing terms energy by PIEDA [kcal/mol]. | DI(MP2)Dispersion energy by PIEDA [kcal/mol]. | q(I=>J)Charge transfer value from I to J fragmens [e]. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D | 4 | ASN | 0 | -0.011 | -0.007 | 15.409 | 1.047 | 1.047 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | D | 5 | SER | 0 | -0.005 | 0.000 | 16.130 | -0.947 | -0.947 | 0.000 | 0.000 | 0.000 | 0.000 |
| 3 | D | 6 | PHE | 0 | -0.003 | 0.005 | 10.302 | 0.462 | 0.462 | 0.000 | 0.000 | 0.000 | 0.000 |
| 4 | D | 7 | SER | 0 | 0.018 | 0.004 | 13.020 | -1.323 | -1.323 | 0.000 | 0.000 | 0.000 | 0.000 |
| 5 | D | 8 | GLY | 0 | 0.018 | 0.002 | 12.303 | 1.673 | 1.673 | 0.000 | 0.000 | 0.000 | 0.000 |
| 6 | D | 9 | TYR | 0 | -0.063 | -0.049 | 8.080 | 1.477 | 1.477 | 0.000 | 0.000 | 0.000 | 0.000 |
| 7 | D | 10 | LEU | 0 | -0.005 | 0.008 | 12.116 | -1.860 | -1.860 | 0.000 | 0.000 | 0.000 | 0.000 |
| 8 | D | 11 | LYS | 1 | 0.840 | 0.900 | 13.637 | -23.794 | -23.794 | 0.000 | 0.000 | 0.000 | 0.000 |
| 9 | D | 12 | LEU | 0 | -0.013 | 0.011 | 12.196 | -0.577 | -0.577 | 0.000 | 0.000 | 0.000 | 0.000 |
| 10 | D | 13 | THR | 0 | 0.041 | 0.007 | 16.471 | -0.996 | -0.996 | 0.000 | 0.000 | 0.000 | 0.000 |
| 11 | D | 14 | ASP | -1 | -0.889 | -0.939 | 18.418 | 24.293 | 24.293 | 0.000 | 0.000 | 0.000 | 0.000 |
| 12 | D | 15 | ASN | 0 | -0.041 | -0.020 | 16.524 | -1.462 | -1.462 | 0.000 | 0.000 | 0.000 | 0.000 |
| 13 | D | 16 | VAL | 0 | 0.029 | 0.024 | 11.976 | 1.850 | 1.850 | 0.000 | 0.000 | 0.000 | 0.000 |
| 14 | D | 17 | TYR | 0 | -0.004 | 0.000 | 12.426 | -2.064 | -2.064 | 0.000 | 0.000 | 0.000 | 0.000 |
| 15 | D | 18 | ILE | 0 | 0.033 | 0.025 | 8.138 | 3.207 | 3.207 | 0.000 | 0.000 | 0.000 | 0.000 |
| 16 | D | 19 | LYS | 1 | 0.849 | 0.905 | 7.576 | -38.639 | -38.639 | 0.000 | 0.000 | 0.000 | 0.000 |
| 17 | D | 20 | ASN | 0 | -0.055 | -0.026 | 6.047 | 0.910 | 0.910 | 0.000 | 0.000 | 0.000 | 0.000 |
| 18 | D | 21 | ALA | 0 | 0.012 | -0.011 | 4.552 | -3.736 | -3.534 | -0.001 | -0.025 | -0.176 | 0.000 |
| 19 | D | 22 | ASP | -1 | -0.751 | -0.877 | 1.737 | 44.788 | 42.156 | 14.253 | -5.944 | -5.677 | -0.049 |
| 20 | D | 23 | ILE | 0 | 0.027 | -0.001 | 2.003 | -17.437 | -19.124 | 11.508 | -3.533 | -6.288 | 0.029 |
| 21 | D | 24 | VAL | 0 | -0.036 | -0.021 | 3.200 | -7.083 | -6.021 | 0.041 | -0.359 | -0.744 | 0.003 |
| 22 | D | 25 | GLU | -1 | -0.859 | -0.920 | 5.093 | 32.428 | 32.428 | 0.000 | 0.000 | 0.000 | 0.000 |
| 23 | D | 26 | GLU | -1 | -0.781 | -0.869 | 6.279 | 33.877 | 33.877 | 0.000 | 0.000 | 0.000 | 0.000 |
| 24 | D | 27 | ALA | 0 | 0.014 | 0.010 | 7.800 | -2.953 | -2.953 | 0.000 | 0.000 | 0.000 | 0.000 |
| 25 | D | 28 | LYS | 1 | 0.869 | 0.936 | 9.538 | -38.185 | -38.185 | 0.000 | 0.000 | 0.000 | 0.000 |
| 26 | D | 29 | LYS | 1 | 0.769 | 0.872 | 9.034 | -37.838 | -37.838 | 0.000 | 0.000 | 0.000 | 0.000 |
| 27 | D | 30 | VAL | 0 | -0.005 | 0.013 | 11.045 | -1.590 | -1.590 | 0.000 | 0.000 | 0.000 | 0.000 |
| 28 | D | 31 | LYS | 1 | 0.836 | 0.930 | 13.695 | -32.494 | -32.494 | 0.000 | 0.000 | 0.000 | 0.000 |
| 29 | D | 32 | PRO | 0 | 0.028 | 0.025 | 11.288 | -1.274 | -1.274 | 0.000 | 0.000 | 0.000 | 0.000 |
| 30 | D | 33 | THR | 0 | 0.012 | -0.011 | 14.524 | -0.647 | -0.647 | 0.000 | 0.000 | 0.000 | 0.000 |
| 31 | D | 34 | VAL | 0 | 0.038 | 0.009 | 13.775 | -0.179 | -0.179 | 0.000 | 0.000 | 0.000 | 0.000 |
| 32 | D | 35 | VAL | 0 | -0.013 | 0.009 | 8.639 | 0.573 | 0.573 | 0.000 | 0.000 | 0.000 | 0.000 |
| 33 | D | 36 | VAL | 0 | -0.013 | -0.004 | 7.800 | -0.946 | -0.946 | 0.000 | 0.000 | 0.000 | 0.000 |
| 34 | D | 37 | ASN | 0 | -0.025 | -0.033 | 4.588 | 13.099 | 13.343 | -0.001 | -0.017 | -0.226 | 0.000 |
| 35 | D | 38 | ALA | 0 | 0.001 | 0.018 | 2.562 | -8.975 | -6.493 | 1.961 | -1.820 | -2.624 | 0.021 |
| 36 | D | 39 | ALA | 0 | 0.015 | -0.006 | 2.392 | -5.083 | -0.848 | 3.431 | -2.923 | -4.744 | -0.038 |
| 37 | D | 40 | ASN | 0 | -0.013 | -0.044 | 1.867 | -13.285 | -14.450 | 11.076 | -4.315 | -5.596 | 0.027 |
| 38 | D | 41 | VAL | 0 | 0.035 | 0.011 | 3.894 | -2.744 | -2.590 | 0.006 | -0.024 | -0.136 | 0.000 |
| 39 | D | 42 | TYR | 0 | 0.008 | 0.004 | 6.559 | -3.852 | -3.852 | 0.000 | 0.000 | 0.000 | 0.000 |
| 40 | D | 43 | LEU | 0 | -0.021 | -0.002 | 5.165 | -1.437 | -1.437 | 0.000 | 0.000 | 0.000 | 0.000 |
| 41 | D | 44 | LYS | 1 | 0.825 | 0.917 | 4.857 | -39.153 | -39.153 | 0.000 | 0.000 | 0.000 | 0.000 |
| 42 | D | 45 | HIS | 0 | -0.008 | -0.022 | 2.752 | 8.804 | 11.530 | 0.907 | -1.611 | -2.023 | -0.002 |
| 43 | D | 46 | GLY | 0 | 0.012 | 0.006 | 1.946 | -9.772 | -11.999 | 6.937 | -1.997 | -2.712 | 0.019 |
| 44 | D | 47 | GLY | 0 | -0.013 | -0.016 | 2.743 | -1.126 | -0.217 | 0.819 | -0.283 | -1.444 | -0.008 |
| 45 | D | 48 | GLY | 0 | 0.033 | 0.028 | 2.324 | -9.548 | -7.656 | 5.477 | -3.321 | -4.048 | 0.029 |
| 46 | D | 49 | VAL | 0 | 0.006 | -0.015 | 1.748 | -37.488 | -40.808 | 18.449 | -4.191 | -10.937 | 0.070 |
| 47 | D | 50 | ALA | 0 | 0.009 | 0.015 | 2.228 | -25.060 | -23.258 | 4.747 | -2.432 | -4.118 | 0.017 |
| 48 | D | 51 | GLY | 0 | 0.049 | 0.029 | 3.530 | -14.717 | -14.103 | 0.011 | -0.192 | -0.432 | 0.002 |
| 49 | D | 52 | ALA | 0 | -0.065 | -0.035 | 3.476 | -11.936 | -11.514 | 0.012 | -0.076 | -0.358 | 0.000 |
| 50 | D | 53 | LEU | 0 | 0.022 | -0.003 | 5.375 | -8.448 | -8.309 | -0.001 | -0.003 | -0.135 | 0.000 |
| 51 | D | 54 | ASN | 0 | 0.052 | 0.016 | 7.491 | -8.215 | -8.215 | 0.000 | 0.000 | 0.000 | 0.000 |
| 52 | D | 55 | LYS | 1 | 0.958 | 0.979 | 7.213 | -49.614 | -49.614 | 0.000 | 0.000 | 0.000 | 0.000 |
| 53 | D | 56 | ALA | 0 | -0.038 | -0.010 | 8.329 | -3.825 | -3.825 | 0.000 | 0.000 | 0.000 | 0.000 |
| 54 | D | 57 | THR | 0 | -0.043 | -0.013 | 10.315 | -2.892 | -2.892 | 0.000 | 0.000 | 0.000 | 0.000 |
| 55 | D | 58 | ASN | 0 | -0.056 | -0.037 | 13.194 | -1.821 | -1.821 | 0.000 | 0.000 | 0.000 | 0.000 |
| 56 | D | 59 | ASN | 0 | 0.019 | 0.015 | 11.318 | 0.592 | 0.592 | 0.000 | 0.000 | 0.000 | 0.000 |
| 57 | D | 60 | ALA | 0 | 0.035 | 0.025 | 12.961 | -0.247 | -0.247 | 0.000 | 0.000 | 0.000 | 0.000 |
| 58 | D | 61 | MET | 0 | 0.025 | 0.019 | 8.724 | 1.305 | 1.305 | 0.000 | 0.000 | 0.000 | 0.000 |
| 59 | D | 62 | GLN | 0 | 0.009 | 0.005 | 6.961 | 3.709 | 3.709 | 0.000 | 0.000 | 0.000 | 0.000 |
| 60 | D | 63 | VAL | 0 | -0.010 | 0.003 | 9.935 | -0.935 | -0.935 | 0.000 | 0.000 | 0.000 | 0.000 |
| 61 | D | 64 | GLU | -1 | -0.757 | -0.871 | 13.064 | 33.542 | 33.542 | 0.000 | 0.000 | 0.000 | 0.000 |
| 62 | D | 65 | SER | 0 | -0.008 | -0.011 | 8.704 | 0.623 | 0.623 | 0.000 | 0.000 | 0.000 | 0.000 |
| 63 | D | 66 | ASP | -1 | -0.813 | -0.882 | 10.272 | 35.974 | 35.974 | 0.000 | 0.000 | 0.000 | 0.000 |
| 64 | D | 67 | ASP | -1 | -0.936 | -0.958 | 12.135 | 25.266 | 25.266 | 0.000 | 0.000 | 0.000 | 0.000 |
| 65 | D | 68 | TYR | 0 | 0.061 | 0.008 | 11.940 | -0.911 | -0.911 | 0.000 | 0.000 | 0.000 | 0.000 |
| 66 | D | 69 | ILE | 0 | -0.060 | -0.029 | 9.775 | -0.443 | -0.443 | 0.000 | 0.000 | 0.000 | 0.000 |
| 67 | D | 70 | ALA | 0 | -0.057 | -0.019 | 13.201 | -0.889 | -0.889 | 0.000 | 0.000 | 0.000 | 0.000 |
| 68 | D | 71 | THR | 0 | -0.046 | -0.012 | 16.368 | -1.429 | -1.429 | 0.000 | 0.000 | 0.000 | 0.000 |
| 69 | D | 72 | ASN | 0 | -0.072 | -0.048 | 15.822 | -0.660 | -0.660 | 0.000 | 0.000 | 0.000 | 0.000 |
| 70 | D | 73 | GLY | 0 | 0.004 | 0.020 | 15.153 | -0.243 | -0.243 | 0.000 | 0.000 | 0.000 | 0.000 |
| 71 | D | 74 | PRO | 0 | -0.051 | -0.036 | 11.355 | 0.978 | 0.978 | 0.000 | 0.000 | 0.000 | 0.000 |
| 72 | D | 75 | LEU | 0 | -0.009 | -0.003 | 8.679 | -1.016 | -1.016 | 0.000 | 0.000 | 0.000 | 0.000 |
| 73 | D | 76 | LYS | 1 | 0.872 | 0.931 | 11.351 | -36.777 | -36.777 | 0.000 | 0.000 | 0.000 | 0.000 |
| 74 | D | 77 | VAL | 0 | 0.070 | 0.025 | 8.706 | 1.739 | 1.739 | 0.000 | 0.000 | 0.000 | 0.000 |
| 75 | D | 78 | GLY | 0 | 0.021 | 0.018 | 9.490 | -3.797 | -3.797 | 0.000 | 0.000 | 0.000 | 0.000 |
| 76 | D | 79 | GLY | 0 | -0.060 | -0.024 | 10.987 | -2.475 | -2.475 | 0.000 | 0.000 | 0.000 | 0.000 |
| 77 | D | 80 | SER | 0 | -0.010 | -0.037 | 10.239 | 3.644 | 3.644 | 0.000 | 0.000 | 0.000 | 0.000 |
| 78 | D | 81 | CYS | 0 | -0.028 | 0.012 | 11.658 | -1.671 | -1.671 | 0.000 | 0.000 | 0.000 | 0.000 |
| 79 | D | 82 | VAL | 0 | 0.011 | 0.008 | 12.472 | 2.528 | 2.528 | 0.000 | 0.000 | 0.000 | 0.000 |
| 80 | D | 83 | LEU | 0 | 0.022 | 0.024 | 10.800 | -1.748 | -1.748 | 0.000 | 0.000 | 0.000 | 0.000 |
| 81 | D | 84 | SER | 0 | 0.009 | -0.001 | 14.514 | -0.032 | -0.032 | 0.000 | 0.000 | 0.000 | 0.000 |
| 82 | D | 85 | GLY | 0 | -0.005 | 0.000 | 13.805 | 2.404 | 2.404 | 0.000 | 0.000 | 0.000 | 0.000 |
| 83 | D | 86 | HIS | 0 | -0.041 | -0.038 | 13.700 | -0.366 | -0.366 | 0.000 | 0.000 | 0.000 | 0.000 |
| 84 | D | 87 | ASN | 0 | -0.030 | -0.029 | 13.242 | 0.082 | 0.082 | 0.000 | 0.000 | 0.000 | 0.000 |
| 85 | D | 88 | LEU | 0 | -0.047 | -0.038 | 8.796 | -0.719 | -0.719 | 0.000 | 0.000 | 0.000 | 0.000 |
| 86 | D | 89 | ALA | 0 | -0.001 | 0.006 | 12.304 | -0.229 | -0.229 | 0.000 | 0.000 | 0.000 | 0.000 |
| 87 | D | 90 | LYS | 1 | 0.916 | 0.985 | 14.539 | -26.187 | -26.187 | 0.000 | 0.000 | 0.000 | 0.000 |
| 88 | D | 91 | HIS | 0 | -0.002 | -0.012 | 16.372 | -1.776 | -1.776 | 0.000 | 0.000 | 0.000 | 0.000 |
| 89 | D | 92 | CYS | 0 | -0.006 | -0.005 | 9.801 | 2.674 | 2.674 | 0.000 | 0.000 | 0.000 | 0.000 |
| 90 | D | 93 | LEU | 0 | -0.045 | -0.001 | 11.949 | -1.336 | -1.336 | 0.000 | 0.000 | 0.000 | 0.000 |
| 91 | D | 94 | HIS | 0 | 0.057 | 0.010 | 7.719 | 8.352 | 8.352 | 0.000 | 0.000 | 0.000 | 0.000 |
| 92 | D | 95 | VAL | 0 | 0.031 | 0.018 | 7.639 | -4.307 | -4.307 | 0.000 | 0.000 | 0.000 | 0.000 |
| 93 | D | 96 | VAL | 0 | -0.035 | 0.004 | 5.467 | 8.320 | 8.320 | 0.000 | 0.000 | 0.000 | 0.000 |
| 94 | D | 97 | GLY | 0 | 0.032 | 0.015 | 4.754 | -6.906 | -6.830 | -0.001 | -0.011 | -0.064 | 0.000 |
| 95 | D | 98 | PRO | 0 | -0.013 | -0.002 | 5.676 | 0.923 | 0.987 | -0.001 | -0.008 | -0.055 | 0.000 |
| 96 | D | 99 | ASN | 0 | 0.008 | 0.005 | 4.523 | 9.157 | 9.400 | -0.001 | -0.015 | -0.226 | 0.000 |
| 97 | D | 100 | VAL | 0 | 0.029 | 0.011 | 7.011 | -7.379 | -7.379 | 0.000 | 0.000 | 0.000 | 0.000 |
| 98 | D | 101 | ASN | 0 | -0.036 | -0.009 | 6.725 | -10.779 | -10.779 | 0.000 | 0.000 | 0.000 | 0.000 |
| 99 | D | 102 | LYS | 1 | 0.870 | 0.937 | 6.531 | -58.425 | -58.425 | 0.000 | 0.000 | 0.000 | 0.000 |
| 100 | D | 103 | GLY | 0 | -0.001 | 0.005 | 10.461 | -2.425 | -2.425 | 0.000 | 0.000 | 0.000 | 0.000 |
| 101 | D | 104 | GLU | -1 | -0.894 | -0.940 | 8.926 | 52.728 | 52.728 | 0.000 | 0.000 | 0.000 | 0.000 |
| 102 | D | 105 | ASP | -1 | -0.829 | -0.920 | 12.024 | 37.609 | 37.609 | 0.000 | 0.000 | 0.000 | 0.000 |
| 103 | D | 106 | ILE | 0 | 0.043 | 0.013 | 9.301 | 2.195 | 2.195 | 0.000 | 0.000 | 0.000 | 0.000 |
| 104 | D | 107 | GLN | 0 | 0.006 | 0.006 | 11.517 | 2.171 | 2.171 | 0.000 | 0.000 | 0.000 | 0.000 |
| 105 | D | 108 | LEU | 0 | -0.026 | -0.019 | 10.926 | -0.517 | -0.517 | 0.000 | 0.000 | 0.000 | 0.000 |
| 106 | D | 109 | LEU | 0 | -0.013 | -0.011 | 6.778 | 1.393 | 1.393 | 0.000 | 0.000 | 0.000 | 0.000 |
| 107 | D | 110 | LYS | 1 | 0.909 | 0.956 | 9.846 | -32.718 | -32.718 | 0.000 | 0.000 | 0.000 | 0.000 |
| 108 | D | 111 | SER | 0 | -0.004 | -0.011 | 12.759 | -1.228 | -1.228 | 0.000 | 0.000 | 0.000 | 0.000 |
| 109 | D | 112 | ALA | 0 | 0.003 | -0.005 | 8.000 | -0.771 | -0.771 | 0.000 | 0.000 | 0.000 | 0.000 |
| 110 | D | 113 | TYR | 0 | 0.038 | 0.002 | 6.492 | 0.790 | 0.790 | 0.000 | 0.000 | 0.000 | 0.000 |
| 111 | D | 114 | GLU | -1 | -0.837 | -0.908 | 11.503 | 30.181 | 30.181 | 0.000 | 0.000 | 0.000 | 0.000 |
| 112 | D | 115 | ASN | 0 | -0.077 | -0.025 | 11.695 | -3.469 | -3.469 | 0.000 | 0.000 | 0.000 | 0.000 |
| 113 | D | 116 | PHE | 0 | 0.025 | -0.013 | 8.252 | -0.445 | -0.445 | 0.000 | 0.000 | 0.000 | 0.000 |
| 114 | D | 117 | ASN | 0 | 0.004 | -0.007 | 14.576 | -0.941 | -0.941 | 0.000 | 0.000 | 0.000 | 0.000 |
| 115 | D | 118 | GLN | 0 | -0.070 | -0.026 | 17.474 | -1.303 | -1.303 | 0.000 | 0.000 | 0.000 | 0.000 |
| 116 | D | 119 | HIS | 0 | -0.002 | -0.006 | 16.413 | -0.961 | -0.961 | 0.000 | 0.000 | 0.000 | 0.000 |
| 117 | D | 120 | GLU | -1 | -0.923 | -0.953 | 18.379 | 25.315 | 25.315 | 0.000 | 0.000 | 0.000 | 0.000 |
| 118 | D | 121 | VAL | 0 | 0.001 | -0.023 | 14.842 | 0.070 | 0.070 | 0.000 | 0.000 | 0.000 | 0.000 |
| 119 | D | 122 | LEU | 0 | -0.060 | -0.011 | 12.668 | 0.694 | 0.694 | 0.000 | 0.000 | 0.000 | 0.000 |
| 120 | D | 123 | LEU | 0 | -0.027 | 0.004 | 6.478 | -0.724 | -0.724 | 0.000 | 0.000 | 0.000 | 0.000 |
| 121 | D | 124 | ALA | 0 | 0.030 | 0.009 | 7.924 | 1.270 | 1.270 | 0.000 | 0.000 | 0.000 | 0.000 |
| 122 | D | 125 | PRO | 0 | 0.008 | 0.023 | 2.890 | -4.647 | -2.378 | 0.331 | -0.701 | -1.899 | 0.006 |
| 123 | D | 126 | LEU | 0 | 0.004 | -0.003 | 2.363 | -9.092 | -6.424 | 0.835 | -0.898 | -2.605 | 0.005 |
| 124 | D | 127 | LEU | 0 | 0.013 | 0.000 | 2.444 | 5.431 | 8.679 | 2.470 | -2.125 | -3.593 | -0.026 |
| 125 | D | 128 | SER | 0 | 0.042 | -0.014 | 2.100 | -24.542 | -24.339 | 8.738 | -4.505 | -4.435 | 0.052 |
| 126 | D | 129 | ALA | 0 | -0.046 | -0.007 | 2.287 | -3.597 | -6.213 | 3.568 | 3.331 | -4.283 | 0.000 |
| 127 | D | 130 | GLY | 0 | 0.015 | 0.003 | 1.840 | -29.162 | -33.139 | 18.200 | -7.295 | -6.929 | 0.041 |
| 128 | D | 131 | ILE | 0 | -0.042 | -0.023 | 1.850 | -36.957 | -39.015 | 13.328 | -1.175 | -10.095 | 0.060 |
| 129 | D | 132 | PHE | 0 | -0.089 | -0.049 | 1.972 | -49.701 | -44.805 | 14.031 | -6.245 | -12.683 | 0.042 |
| 130 | D | 133 | GLY | 0 | 0.040 | 0.024 | 3.787 | -17.679 | -17.264 | 0.015 | -0.033 | -0.398 | 0.002 |
| 131 | D | 134 | ALA | 0 | -0.014 | -0.008 | 4.851 | -12.232 | -12.155 | -0.001 | -0.008 | -0.069 | 0.000 |
| 132 | D | 135 | ASP | -1 | -0.789 | -0.918 | 6.093 | 51.517 | 51.517 | 0.000 | 0.000 | 0.000 | 0.000 |
| 133 | D | 136 | PRO | 0 | -0.008 | -0.010 | 3.634 | -1.471 | -0.860 | 0.012 | -0.186 | -0.438 | 0.001 |
| 134 | D | 137 | ILE | 0 | 0.017 | 0.009 | 5.522 | -0.843 | -0.782 | -0.001 | -0.001 | -0.059 | 0.000 |
| 135 | D | 138 | HIS | 0 | 0.013 | 0.008 | 8.915 | -3.152 | -3.152 | 0.000 | 0.000 | 0.000 | 0.000 |
| 136 | D | 139 | SER | 0 | -0.017 | -0.021 | 5.760 | -4.084 | -4.084 | 0.000 | 0.000 | 0.000 | 0.000 |
| 137 | D | 140 | LEU | 0 | 0.019 | 0.002 | 7.895 | -1.130 | -1.130 | 0.000 | 0.000 | 0.000 | 0.000 |
| 138 | D | 141 | ARG | 1 | 0.908 | 0.973 | 8.870 | -35.107 | -35.107 | 0.000 | 0.000 | 0.000 | 0.000 |
| 139 | D | 142 | VAL | 0 | 0.012 | 0.010 | 10.943 | -2.183 | -2.183 | 0.000 | 0.000 | 0.000 | 0.000 |
| 140 | D | 143 | CYS | 0 | -0.048 | 0.009 | 9.061 | -2.318 | -2.318 | 0.000 | 0.000 | 0.000 | 0.000 |
| 141 | D | 144 | VAL | 0 | 0.003 | 0.000 | 11.218 | -2.353 | -2.353 | 0.000 | 0.000 | 0.000 | 0.000 |
| 142 | D | 145 | ASP | -1 | -0.876 | -0.947 | 13.971 | 29.983 | 29.983 | 0.000 | 0.000 | 0.000 | 0.000 |
| 143 | D | 146 | THR | 0 | -0.145 | -0.091 | 14.200 | -2.306 | -2.306 | 0.000 | 0.000 | 0.000 | 0.000 |
| 144 | D | 147 | VAL | 0 | -0.027 | 0.014 | 12.369 | -0.808 | -0.808 | 0.000 | 0.000 | 0.000 | 0.000 |
| 145 | D | 148 | ARG | 1 | 0.827 | 0.905 | 15.650 | -30.966 | -30.966 | 0.000 | 0.000 | 0.000 | 0.000 |
| 146 | D | 149 | THR | 0 | -0.006 | 0.021 | 17.347 | -1.005 | -1.005 | 0.000 | 0.000 | 0.000 | 0.000 |
| 147 | D | 150 | ASN | 0 | -0.037 | -0.010 | 16.494 | 0.691 | 0.691 | 0.000 | 0.000 | 0.000 | 0.000 |
| 148 | D | 151 | VAL | 0 | 0.033 | 0.015 | 10.800 | 1.118 | 1.118 | 0.000 | 0.000 | 0.000 | 0.000 |
| 149 | D | 152 | TYR | 0 | -0.035 | -0.022 | 9.995 | -1.274 | -1.274 | 0.000 | 0.000 | 0.000 | 0.000 |
| 150 | D | 153 | LEU | 0 | 0.014 | -0.009 | 6.988 | 2.860 | 2.860 | 0.000 | 0.000 | 0.000 | 0.000 |
| 151 | D | 154 | ALA | 0 | -0.017 | -0.009 | 3.111 | -4.390 | -3.803 | 0.043 | -0.146 | -0.484 | 0.000 |
| 152 | D | 155 | VAL | 0 | 0.016 | 0.009 | 2.687 | 7.868 | 10.656 | 1.242 | -1.438 | -2.591 | -0.010 |
| 153 | D | 156 | PHE | 0 | 0.021 | 0.010 | 2.591 | -19.274 | -12.755 | 6.870 | -3.740 | -9.650 | 0.037 |
| 154 | D | 157 | ASP | -1 | -0.840 | -0.901 | 3.902 | 39.908 | 40.409 | 0.026 | -0.060 | -0.467 | 0.001 |
| 155 | D | 158 | LYS | 1 | 0.871 | 0.910 | 6.477 | -33.255 | -33.255 | 0.000 | 0.000 | 0.000 | 0.000 |
| 156 | D | 159 | ASN | 0 | 0.002 | -0.016 | 9.151 | -3.982 | -3.982 | 0.000 | 0.000 | 0.000 | 0.000 |
| 157 | D | 160 | LEU | 0 | -0.030 | -0.012 | 2.668 | -2.532 | -1.403 | 0.442 | -0.426 | -1.145 | 0.002 |
| 158 | D | 161 | TYR | 0 | -0.002 | 0.002 | 6.789 | -1.094 | -1.094 | 0.000 | 0.000 | 0.000 | 0.000 |
| 159 | D | 162 | ASP | -1 | -0.786 | -0.873 | 8.453 | 31.600 | 31.600 | 0.000 | 0.000 | 0.000 | 0.000 |
| 160 | D | 163 | LYS | 1 | 0.901 | 0.977 | 7.544 | -46.644 | -46.644 | 0.000 | 0.000 | 0.000 | 0.000 |
| 161 | D | 164 | LEU | 0 | -0.053 | -0.023 | 5.142 | -0.681 | -0.681 | 0.000 | 0.000 | 0.000 | 0.000 |
| 162 | D | 165 | VAL | 0 | 0.007 | -0.007 | 9.545 | -1.468 | -1.468 | 0.000 | 0.000 | 0.000 | 0.000 |
| 163 | D | 166 | SER | 0 | -0.056 | -0.031 | 12.767 | -1.922 | -1.922 | 0.000 | 0.000 | 0.000 | 0.000 |
| 164 | D | 167 | SER | 0 | -0.091 | -0.060 | 11.894 | -0.936 | -0.936 | 0.000 | 0.000 | 0.000 | 0.000 |
| 165 | D | 168 | PHE | -1 | -0.914 | -0.950 | 13.126 | 29.068 | 29.068 | 0.000 | 0.000 | 0.000 | 0.000 |